Research
Our interests lie at the interface between theoretical/computational chemistry and biophysics. The current research in the lab is directed towards understanding how biomolecules perform their functions via dynamical motions that are encoded in their three dimensional structures. Ongoing projects include computer simulations of ABC-transporters and development of combined quantum mechanical/molecular mechanical (QM/MM) methods.
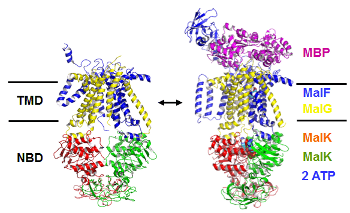
ABC-transporters
Many essential functions of living cells are performed by nanoscale motors
consisting of protein complexes. The ability of these biomolecular motors to
utilize chemical free energy to perform mechanical work makes them splendid
molecular machines. Among various types of molecular motors, ATP-binding cassette
(ABC) transporters represent a unique family of motor proteins that enable translocations
of various substrates across cell membranes, by harnessing the free energy associated with ATP
binding and hydrolysis.[1] Dysfunctions of ABC-transporters have been linked to a number
of diseases, including cystic fibrosis, the most common fatal hereditary disease in the US.
Over expressions of certain ABC-transporters are also known to contribute to multidrug
resistance of tumor cells after cancer patients receive chemotherapy. Making using multiscale computer simulations,
our project aims at obtaining a deeper understanding of conformational dynamics, enzyme catalysis, as well as the chemomechanical coupling mechanisms under which the chemical free energy is converted into mechanical work in ABC-transporters. Currently, we are focusing on the elucidation of ATP hydrolysis mechanisms in several ABC systems.
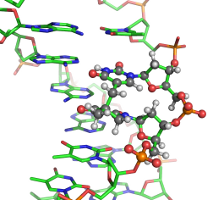
DNA damages
In scarce of nutrients, certain bacteria form endospores, which protect bacterial DNAs from various environmental damaging factors, including UV radiation. It is well known that UV radiation can damage DNAs by forming cross-links between adjacent Thymine bases (TT) in a DNA strand. Under regular cellular conditions, such UV-induced damage leads to the formation of cyclobutane pyrimidine dimer (CPD) as the dominant photoproduct. In spores, however, the DNA photoreaction pattern is altered such that a special lesion 5-thyminyl-5-6-dihydrothymine, or commonly known as the spore-photoproduct (SP), becomes the most prevalent form of TT damages. It has been postulated that SP lesions are formed in a well-controlled manner such that the bacterial DNAs are "locked" at a chemically inert state until spores progress to the germination phase, during which these lesions are repaired faithfully by a specialized enzyme called spore-photoproduct lyase (SPL). Despite this widely accepted overall scenario, the precise mechanisms of how SP is promoted to a major photoproduct in spores and how the SP lesions are recognized by SPL for repair remain largely unknown. Collaborating with Prof. Lei Li, we have computationally characterized several dinucleotide analogues of SP[2][3] and are currently developing all-atom models for simulating short segments of SP-containing DNAs.
QM/MM development
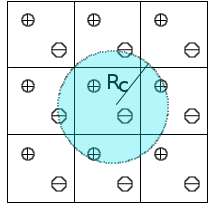
We are exploring new approaches to include long-range electrostatic interactions in combined quantum mechanical and
molecular mechanical (QM/MM) calculations. Traditionally, long-range electrostatics in molecular
simulations can be treated by lattice sum methods such as the Ewald method, where Coulomb interactions are collected over periodic systems such that contributions from all particles as well as their periodic images are taken into account without making any truncation. The Isotropic Periodic Sum (IPS) method[4][5][6] has been developed as an efficient alternative to the Ewald method. In the IPS method, pairwise interactions are only computed in a local region defined by a cutoff distance, where the long range contribution beyond the cutoff distance is treated with a mean field theory by assuming an isotropic distribution of the image particles. To date, the IPS method has been formulated and applied to classical systems that are described by pure molecular mechanical force fields. We are currently developing computer codes to extend the IPS method to treat systems described by combined QM/MM potentials. This work is expected to lay the foundation of applying the IPS method to enzyme simulations.
References
- ↑ Structure, Function, and Evolution of Bacterial ATP-Binding Cassette Systems," Davidson, A. L; Dassa, E.; Orelle, C.; Chen, J. Microbiol. Mol. Biol. Revs. 2008,72 317-364.
- ↑ "Expanding the Horizon of the Thymine Isostere Biochemistry: Unique Dimers formed via Photoreaction between a Thymine and a Toluene residue in the Dinucleotide Framework," Lin, D.; Zhou, Y.; Pu, J.; Li, L. Chem. Eur. J. 2012, 18, 7823-7833.
- ↑ "Chemical Synthesis, Crystal Structure and Enzymatic Evaluation of a Dinucleotide Spore Photoproduct Analogue Containing Formacetal Linker," Lin, G.; Chen, C.-H; Pink, M.; Pu, J.; Li, L. Chem. Eur. J. 2011, 17, 9658-9668.
- ↑ "Isotropic Periodic Sum: a Method for the Calculation of Long-Range Interactions," Wu, X.; Brooks, B. R. J. Chem. Phys. 2005, 122, 44107.
- ↑ "Using the Isotropic Periodic Sum Method to Calculate Long-Range Interactions of Heterogeneous Systems," Wu, X.; Brooks, B. R. J. Chem. Phys. 2008, 129, 154115.
- ↑ "Isotropic Periodic Sum of Electrostatic Interactions for Polar Systems," Wu, X.; Brooks, B. R. J. Chem. Phys. 2009, 131, 024107.